
[ad_1]
Cells depend on complicated molecular machines composed of protein assemblies to carry out important features similar to power manufacturing, gene expression, and protein synthesis. To higher perceive how these machines work, scientists seize snapshots of them by isolating proteins from cells and utilizing numerous strategies to find out their buildings. Nonetheless, isolating proteins from cells additionally removes them from the context of their native surroundings, together with protein interplay companions and mobile location.
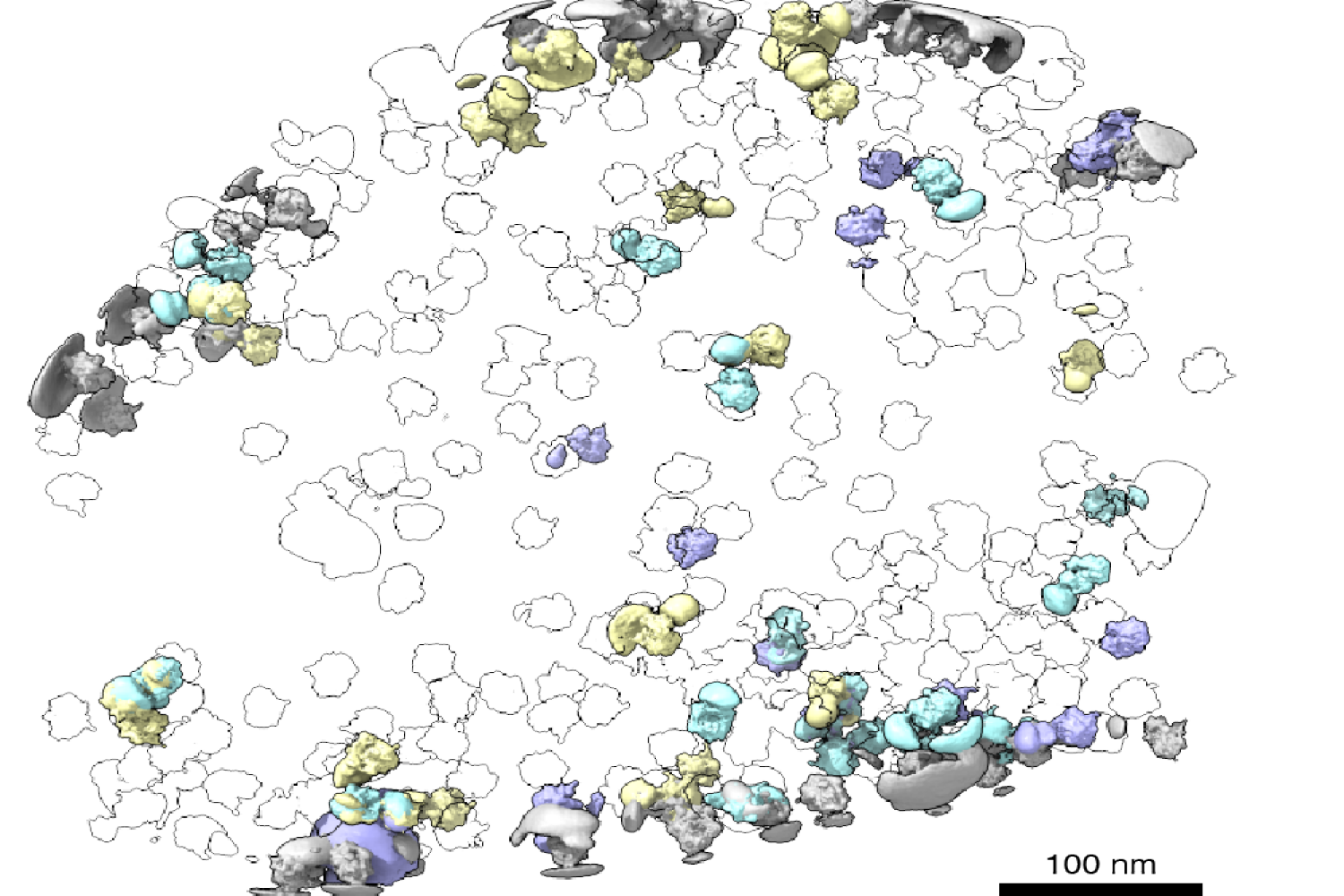
Just lately, cryogenic electron tomography (cryo-ET) has emerged as a approach to observe proteins of their native surroundings by imaging frozen cells at totally different angles to acquire three-dimensional structural info. This method is thrilling as a result of it permits researchers to immediately observe how and the place proteins affiliate with one another, revealing the mobile neighborhood of these interactions throughout the cell.
With the know-how out there to picture proteins of their native surroundings, MIT graduate pupil Barrett Powell puzzled if he may take it one step additional: What if molecular machines might be noticed in motion? In a paper printed March 8 in Nature Strategies, Powell describes the strategy he developed, referred to as tomoDRGN, for modeling structural variations of proteins in cryo-ET knowledge that come up from protein motions or proteins binding to totally different interplay companions. These variations are often known as structural heterogeneity.
Though Powell had joined the lab of MIT affiliate professor of biology Joey Davis as an experimental scientist, he acknowledged the potential affect of computational approaches in understanding structural heterogeneity inside a cell. Beforehand, the Davis Lab developed a associated methodology named cryoDRGN to know structural heterogeneity in purified samples. As Powell and Davis noticed cryo-ET rising in prominence within the area, Powell took on the problem of re-imagining this framework to work in cells.
When fixing buildings with purified samples, every particle is imaged solely as soon as. Against this, cryo-ET knowledge is collected by imaging every particle greater than 40 instances from totally different angles. That meant tomoDRGN wanted to have the ability to merge the knowledge from greater than 40 pictures, which was the place the mission hit a roadblock: the quantity of information led to an info overload.
To deal with this, Powell efficiently rebuilt the cryoDRGN mannequin to prioritize solely the highest-quality knowledge. When imaging the identical particle a number of instances, radiation harm happens. The pictures acquired earlier, due to this fact, are usually of upper high quality as a result of the particles are much less broken.
“By excluding among the lower-quality knowledge, the outcomes have been truly higher than utilizing all the knowledge — and the computational efficiency was considerably quicker,” Powell says.
Simply as Powell was starting work on testing his mannequin, he had a stroke of luck: The authors of a groundbreaking new examine that visualized, for the primary time, ribosomes inside cells at near-atomic decision, shared their uncooked knowledge on the Electrical Microscopy Public Picture Archive (EMPIAR). This dataset was an exemplary check case for Powell, by which he demonstrated that tomoDRGN may uncover structural heterogeneity inside cryo-ET knowledge.
Based on Powell, one thrilling result’s what tomoDRGN discovered surrounding a subset of ribosomes within the EMPIAR dataset. Among the ribosomal particles have been related to a bacterial cell membrane and engaged in a course of referred to as cotranslational translocation. This happens when a protein is being concurrently synthesized and transported throughout a membrane. Researchers can use this end result to make new hypotheses about how the ribosome features with different protein equipment integral to transporting proteins outdoors of the cell, now guided by a construction of the complicated in its native surroundings.
After seeing that tomoDRGN may resolve structural heterogeneity from a structurally numerous dataset, Powell was curious: How small of a inhabitants may tomoDRGN establish? For that check, he selected a protein named apoferritin, which is a generally used benchmark for cryo-ET and is usually handled as structurally homogeneous. Ferritin is a protein used for iron storage and is known as apoferritin when it lacks iron.
Surprisingly, along with the anticipated particles, tomoDRGN revealed a minor inhabitants of ferritin particles — with iron certain — making up simply 2 p.c of the dataset, that was not beforehand reported. This end result additional demonstrated tomoDRGN’s capability to establish structural states that happen so occasionally that they’d be averaged out of a 3D reconstruction.
Powell and different members of the Davis Lab are excited to see how tomoDRGN will be utilized to additional ribosomal research and to different techniques. Davis works on understanding how cells assemble, regulate, and degrade molecular machines, so the following steps embrace exploring ribosome biogenesis inside cells in larger element utilizing this new instrument.
“What are the potential states that we could also be dropping throughout purification?” Davis asks. “Maybe extra excitingly, we are able to take a look at how they localize throughout the cell and what companions and protein complexes they could be interacting with.”
[ad_2]